Aligning two structures
Oftentimes you will want to compare two different PDB files to one another. Here's how.
You can either download the two files that are of interest, and open
them into PyMOL, or you can use the Fetch command if you know the ID's
already. PyMOL will then download the files from the PDB site
remotely. For example, I will compare two structures of the DNA-binding
protein MntR that have PDB ID's 1ON1 and 1ON2.
Open PyMOL and in the command line, type
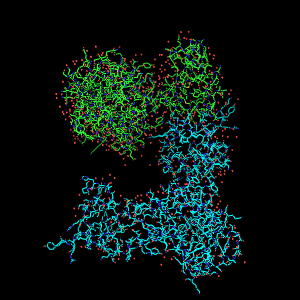
What you'll see is two protein models (labeled 1ON1 and 1ON2 in the Panel).
The simplest command to align them is
align 1ON2, 1ON1
Not that 1ON2 moves while 1ON1 is the fixed reference model. If you
look in the text window, the quality of the alignment isn't that great
(rmsd 0.759 Å). To improve it you can select a subset of residues and
atoms. For example, I know that residues 80-120 form an invariant
structural core to the protein, so I might type:
align 1ON2 and resi 80-120 and name CA, 1ON1
Voila, 0.226 Å rmsd. Next to "all" in the Names Panel, Hide Lines and Show Cartoon, and then Action Zoom to center the image.
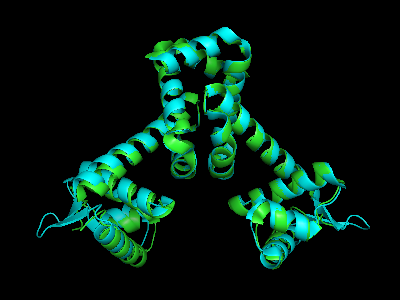
This is a start. For prettier rendering, go here.


